MP-Ref: Reference Genome based Multiplex PCR Primer Design for Versatile Applications #
https://m4.igenetech.com/mpref/
Introduction #
MP-Ref is a reference genome-based server for designing multiplex PCR primers based on Graph-Expanding algorithm. The algorithm performs exhaustive pairwise specificity and dimer analyses at each expansion step to ensure primer compatibility within a single reaction. With over 100 pre-loaded genomes and an intuitive user interface, MP-Ref enables streamlined primer design for versatile applications:
- target enrichment panels for simultaneous amplification of multiple coding gene regions, compatible with short-read or long-read sequencing platforms;
- primer design for short tandem repeat regions by selecting primers flanking the target;
- multiplex primer design with varied amplicon sizes for capillary electrophoresis.
Availability #
The web server is freely available at https://m4.igenetech.com/mpref/.
Demos #
- Singleplex Primer Design with MP-Ref
- Cross-species Specific Primer Design with MP-Ref
- Simutaneously Amplify Two or More Genomes with MP-Ref
- Multiplex PCR Primer Design With Varied Sizes using MP-Ref
- Multiplex PCR Primer Design for Long Target Regions with MP-Ref
- Multiplex PCR Primer Design for SMN1 Whole Region with MP-Ref
Tips #
Designing Primers for Long-Read Sequencing with Large Product Size #
For instance, when designing primers with a target product size of 5,000 bp, it is recommended to set the “Product Min Size” as 4900 and the “Product Max Size” as 5100. Choosing a smaller range such as 4900-5100 is more efficient in terms of computational resources and memory compared to a broader range like 4000-6000. Setting a wider range could potentially strain computational resources and even necessitate server restarts to prevent memory-related issues in extreme cases.
What is the BED format? #
The BED (Browser Extensible Data) format is a text file format used to store genomic regions as coordinates and associated annotations. We just need the first three columns:
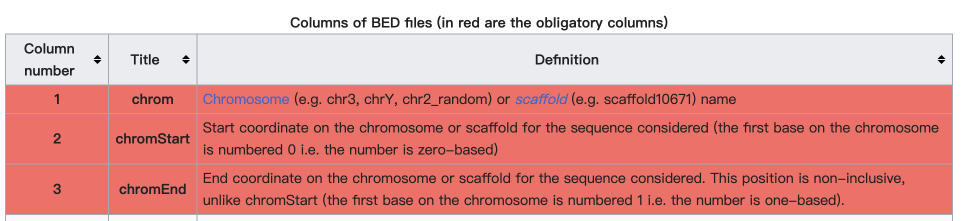 https://en.wikipedia.org/wiki/BED_(file_format)#Description
https://en.wikipedia.org/wiki/BED_(file_format)#Description
Why we need BED format as targets? Why not just paste sequences? #
In addition to identifying the target sequences, it’s crucial to consider the presence of background DNA—DNA other than the intended targets. When designing primers, each pair is evaluated against the entire genome to minimize the likelihood of non-specific amplification. The BED format plays a key role during this evaluation by distinguishing between target and non-target predicted amplicons. However, differentiating them solely based on input sequences poses a challenge for the algorithm.
How to make the targets in BED format? #
To create BED format targets from different input types, follow these protocols:
- For Reference SNPs (rsIDs): rs2bed protocol
- For Gene Symbols: gene2bed protocol
Request more input? #
Designing multiplex PCR primers is a meticulous process, involving genome-wide prediction of nonspecific amplicons for each primer pair within a reaction. Typically, the process takes less than 10 minutes for up to 100 SNPs, with additional time required when incorporating extra background databases. A private link will be created for later access when the task requires a long running time.
To preserve computational resources and prevent misuse, we’ve implemented a cap on the number of targets. Researchers with needs exceeding these limits are encouraged to contact the author. We are more than willing to accommodate your design tasks efficiently.
What’s the target database? And what’s the background databases? How to choose? #
The target database is where the BED-formatted target information originates. Therefore, the chromosome names in the BED format must match those in the genome database.
Background databases are crucial for ensuring that primers do not amplify any unintended sequences from these databases. For instance, in forensic science, it’s essential that a primer pair specifically amplifies the target STR from human genomic DNA, without producing any amplicons from environmental species such as dogs, pigs, or E. coli.
However, the specific needs of researchers can vary across different applications. For example, some researchers might require primers that can simultaneously amplify targets from both genome A and genome B. An example of this is a primer pair designed for human sex identification targeting the AMELX gene:
CCCTGGGCTCTGTAAAGAATAGTG
ATCAGAGCTTAAACTGGGAAGCTG
Result link: https://m4.igenetech.com/spec/demo/e1cb417f-6a71-4a06-9fcb-913848ebd900
How to set “Number of Off-targets”? #
In theory, the ideal setting for this parameter is zero, since the goal is to design primer pairs that do not produce non-target amplicons. However, for certain targets, it’s impractical to achieve such specificity. In these cases, setting the parameter to 10 may be necessary. It’s crucial, though, to ensure that there’s a significant base difference (option “Minimum Nucleotide Differences”) between the target and non-target amplicons, allowing them to be distinguished by next-generation sequencing or other analytical methods.
How to set “Amplicon Count on Background Genomes”? #
In most cases, it’s advisable to set this parameter to zero. A value greater than zero should be considered based on the specific requirements of the application.
What’s the “SNP Skip Size” or “InDel Skip Size”? #
SNPs and InDels located within primer binding sites, especially at the 3’ end, can significantly impact the stability of primer-target binding and negatively affect amplification efficiency. Ideally, primers without SNPs are preferred to ensure high amplification efficiency. However, in regions with high variability and numerous SNPs, finding SNP-free primers may be challenging. In such scenarios, one approach is to lower the SNP tolerance parameter to a value like 6 and attempt the primer design process again. Alternatively, using the “STR” mode to amplify the entire highly variable region might be a viable solution.
Currently, SNP databases configured for primer design are available only for the human genome (versions hg39 and hg19) and the mouse genome (mm10). These common SNP databases are sourced from the UCSC website.
What’s the “Validation method”? #
To validate PCR amplicons, commonly employed methods include Sanger sequencing, next-generation sequencing (NGS), and gel electrophoresis. Each validation method may necessitate a distinct design strategy for the primers. Therefore, selecting an appropriate validation method tailored to the specific requirements of your application is crucial before proceeding with primer design.
When to set “Product Min Difference”? #
When the Validation method is set to size discrimination, this configuration aims to design primers that produce amplicons of varied sizes. These size differences enable the discrimination of specific amplicons using size-selective methods such as gel electrophoresis, facilitating the identification and analysis of target sequences based on their length.
What’s “Dimer ΔG”? #
We use this value to filter out the non-significant (those ΔG > this value) dimers.
Waht’s “Hairpin Score”? Why hairpin uses score and dimer uses ΔG? #
We utilize this score threshold to eliminate non-significant hairpins, those with scores below this value. In our experience, using a score-based approach for hairpin filtering has proven to be efficient.
What’s “Non-specific Amplicon Tm”? #
This parameter is utilized to eliminate nonspecific amplicons with lower melting temperatures (Tm), as derived from MFEprimer.
What’s “Non-specific Amplicon Size (bp)”? #
This range is employed to predict nonspecific amplicons, utilizing parameters from MFEprimer.