How To Get Chromosome Position Given Rs Number?
February 27, 2021
Introduction #
We have a list of SNP rs numbers, like:
rs1002315756
rs1003815568
rs1004109382
rs1004635980
rs1008829651
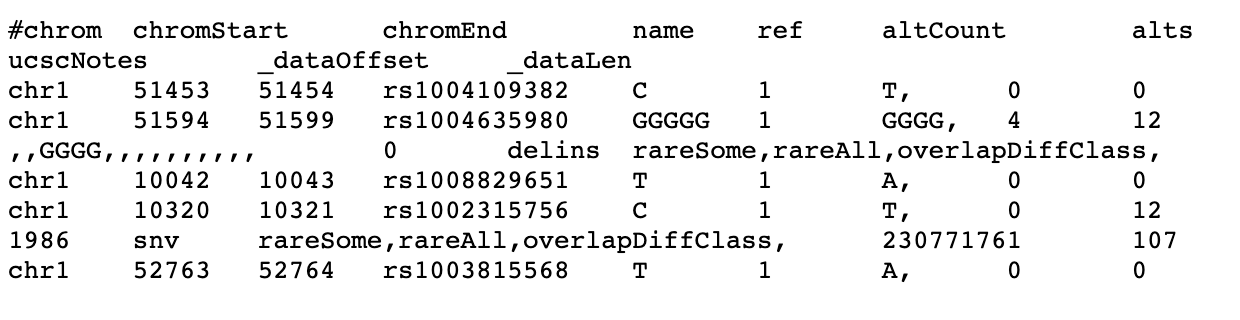
And we want the genome positions for each SNPS, the results should like:
#chrom chromStart chromEnd name ref altCount alts shiftBases freqSourceCount minorAlleleFreq majorAllele minorAllele maxFuncImpact class ucscNotes _dataOffset _dataLen
chr1 51453 51454 rs1004109382 C 1 T, 0 0 0 snv rareSome,rareAll, 409400514 36
chr1 51594 51599 rs1004635980 GGGGG 1 GGGG, 4 12 -inf,-inf,0.000127421,-inf,-inf,-inf,-inf,-inf,-inf,-inf,-inf,-inf, ,,GGGGG,,,,,,,,,, ,,GGGG,,,,,,,,,, 0 delins rareSome,rareAll,overlapDiffClass, 461755619 105
chr1 10042 10043 rs1008829651 T 1 A, 0 0 1986 snv rareSome,rareAll, 878656298 46
chr1 10320 10321 rs1002315756 C 1 T, 0 12 -inf,-inf,-inf,-inf,-inf,0.00102617,-inf,-inf,-inf,-inf,-inf,-inf, ,,,,,C,,,,,,, ,,,,,T,,,,,,, 1986 snv rareSome,rareAll,overlapDiffClass, 230771761 107
chr1 52763 52764 rs1003815568 T 1 A, 0 0 0 snv rareSome,rareAll, 380161821 36
Method #
UCSC Table Browser do this job perfectly, thanks.
- Visit: https://genome.ucsc.edu/cgi-bin/hgTables.
- On the “Table Browser” page, select proper parameters, as “Step 1” in the following picture shows.
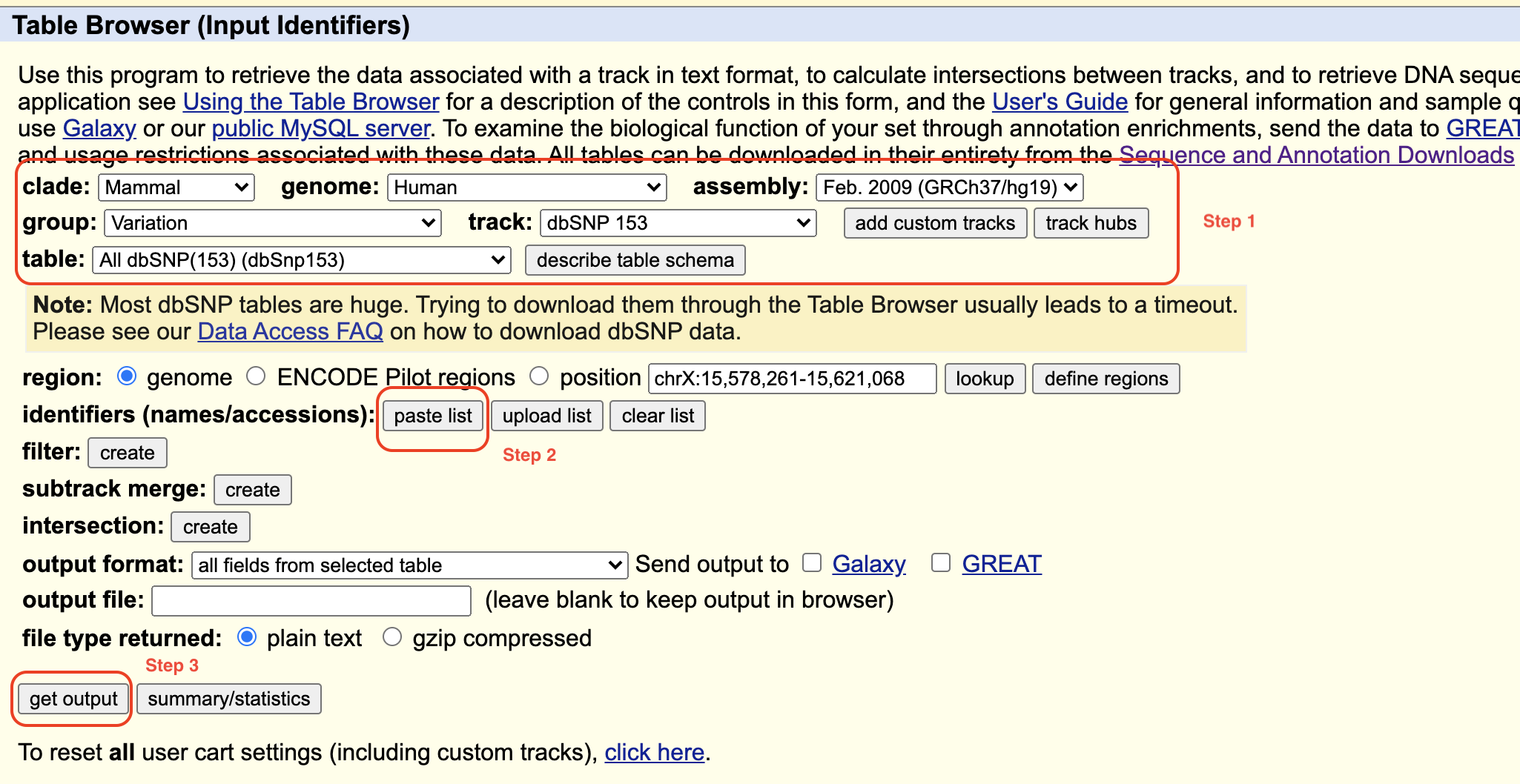
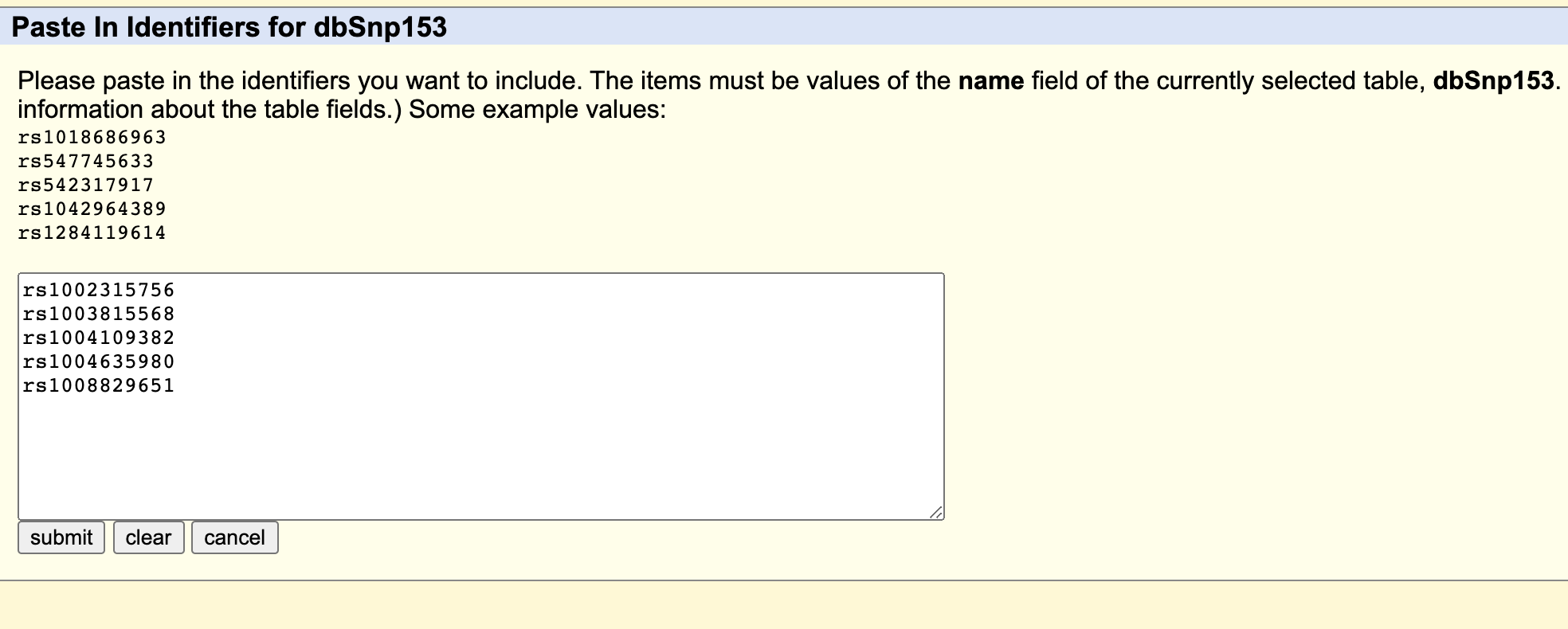
- Click “paste list” in red box labeled as “Step 2”, paste your rs number list as following picture shows, , then click “submit”.
- Click “get output” labeled as “Step 3”.
- We get the genome positions as well as other information, done.