How to get exon coordinates in BED format?
December 20, 2023
Introduction #
We have a list of genes, like:
EGFR
MET
TP53
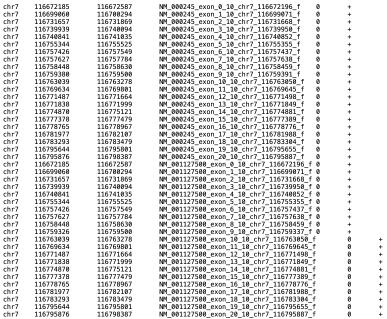
And we want the genome positions plus 10 bases at each end for each exons, the results should like:
chr7 116672185 116672587 NM_000245_exon_0_10_chr7_116672196_f 0 +
chr7 116699060 116700294 NM_000245_exon_1_10_chr7_116699071_f 0 +
chr7 116731657 116731869 NM_000245_exon_2_10_chr7_116731668_f 0 +
chr7 116739939 116740094 NM_000245_exon_3_10_chr7_116739950_f 0 +
chr7 116740841 116741035 NM_000245_exon_4_10_chr7_116740852_f 0 +
chr7 116755344 116755525 NM_000245_exon_5_10_chr7_116755355_f 0 +
chr7 116757426 116757549 NM_000245_exon_6_10_chr7_116757437_f 0 +
chr7 116757627 116757784 NM_000245_exon_7_10_chr7_116757638_f 0 +
chr7 116758448 116758630 NM_000245_exon_8_10_chr7_116758459_f 0 +
chr7 116759380 116759500 NM_000245_exon_9_10_chr7_116759391_f 0 +
Method #
UCSC Table Browser do this job perfectly, thanks.
- Visit: https://genome.ucsc.edu/cgi-bin/hgTables.
- On the “Table Browser” page, select proper parameters.
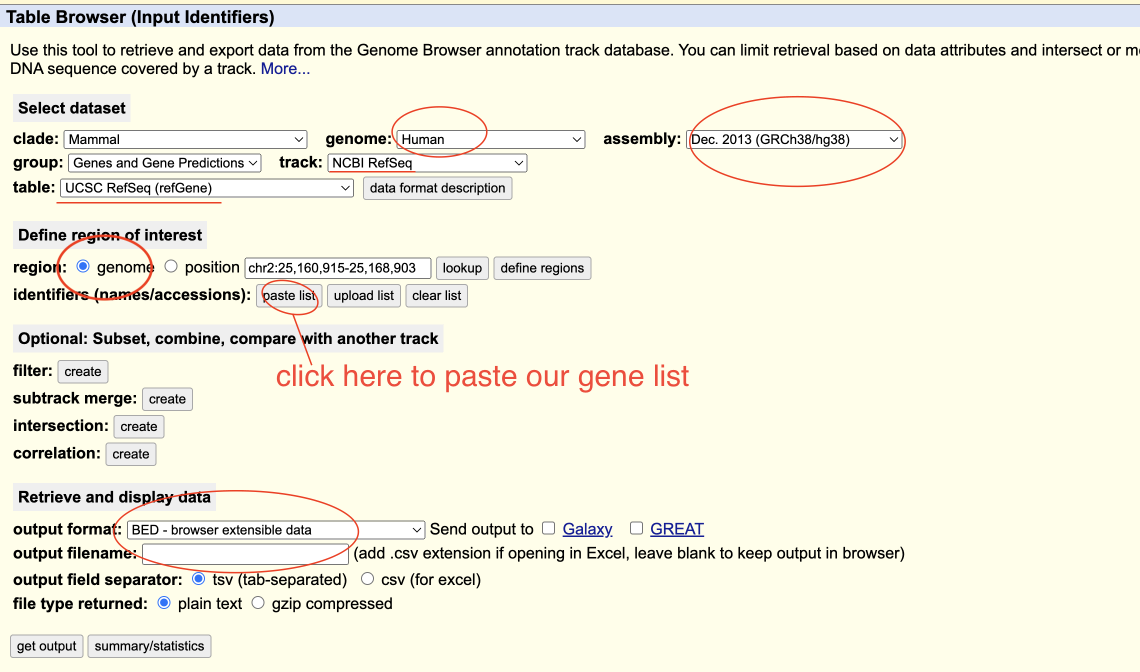
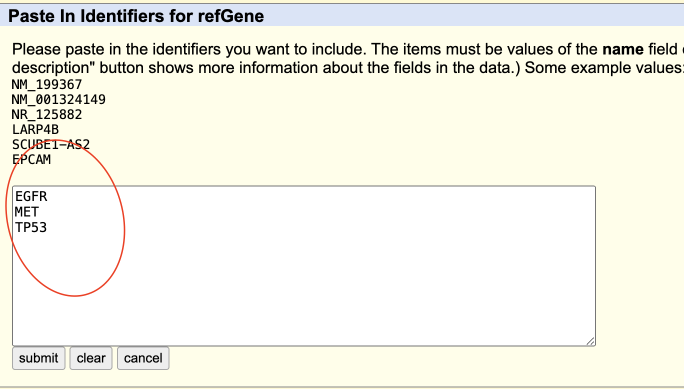
- Click “paste list” and paste gene list as following picture shows, , then click “submit”.
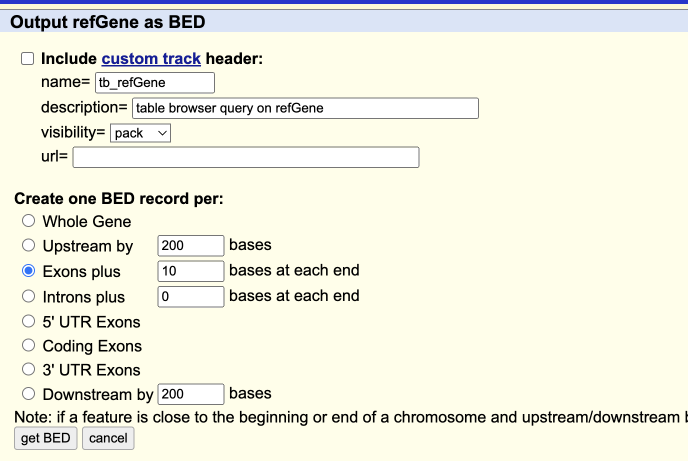
- Select “Exons” plus “10” bases, then click “get BED”
- We get the genome positions as expected, done.