Simutaneously Amplify Two or More Genomes with MP-Ref
July 21, 2025
Designing primers capable of amplifying both genome A and genome B simultaneously using MP-Ref (demo link)。
-
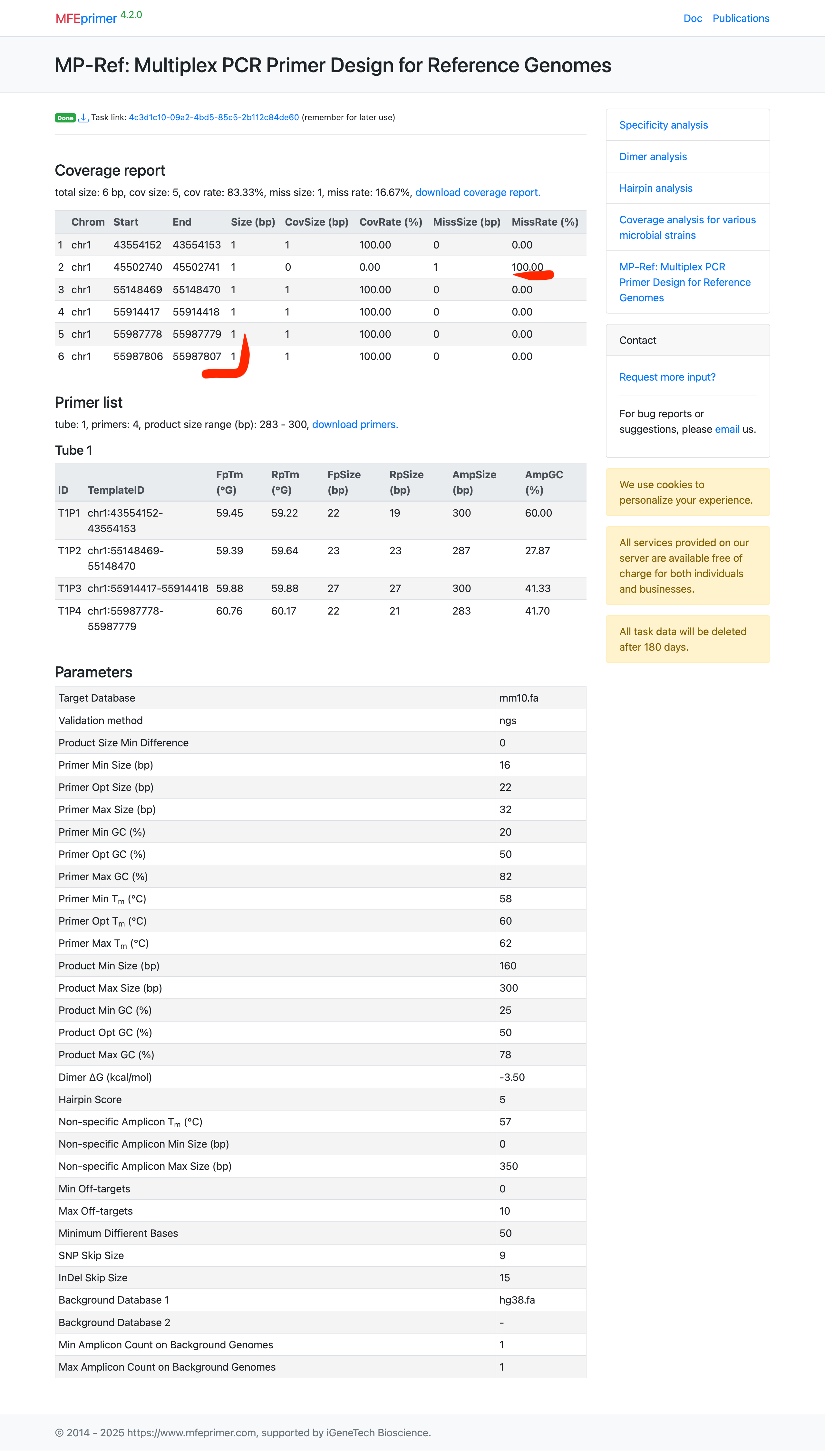
Select the target database: this time we design primers for mouse and requires these primers should also have exactly one amplicon in human genome. Please be noted that we also set “Min nonspecific amplicon Tm” = 57, a bigger higher Tm. This is to tell the server we need a good (with Tm close to the target amplicon in mouse genome) amplicon for human genome.
-
Enter out targets in BED format;
chr1 43554152 43554153 chr1 45502740 45502741 chr1 55148469 55148470 chr1 55914417 55914418 chr1 55987778 55987779 chr1 55987806 55987807 -
Also select backgrounds databases.
-
And set “Min=1 Max=1” for “Amplicon Count on Background Genomes”.
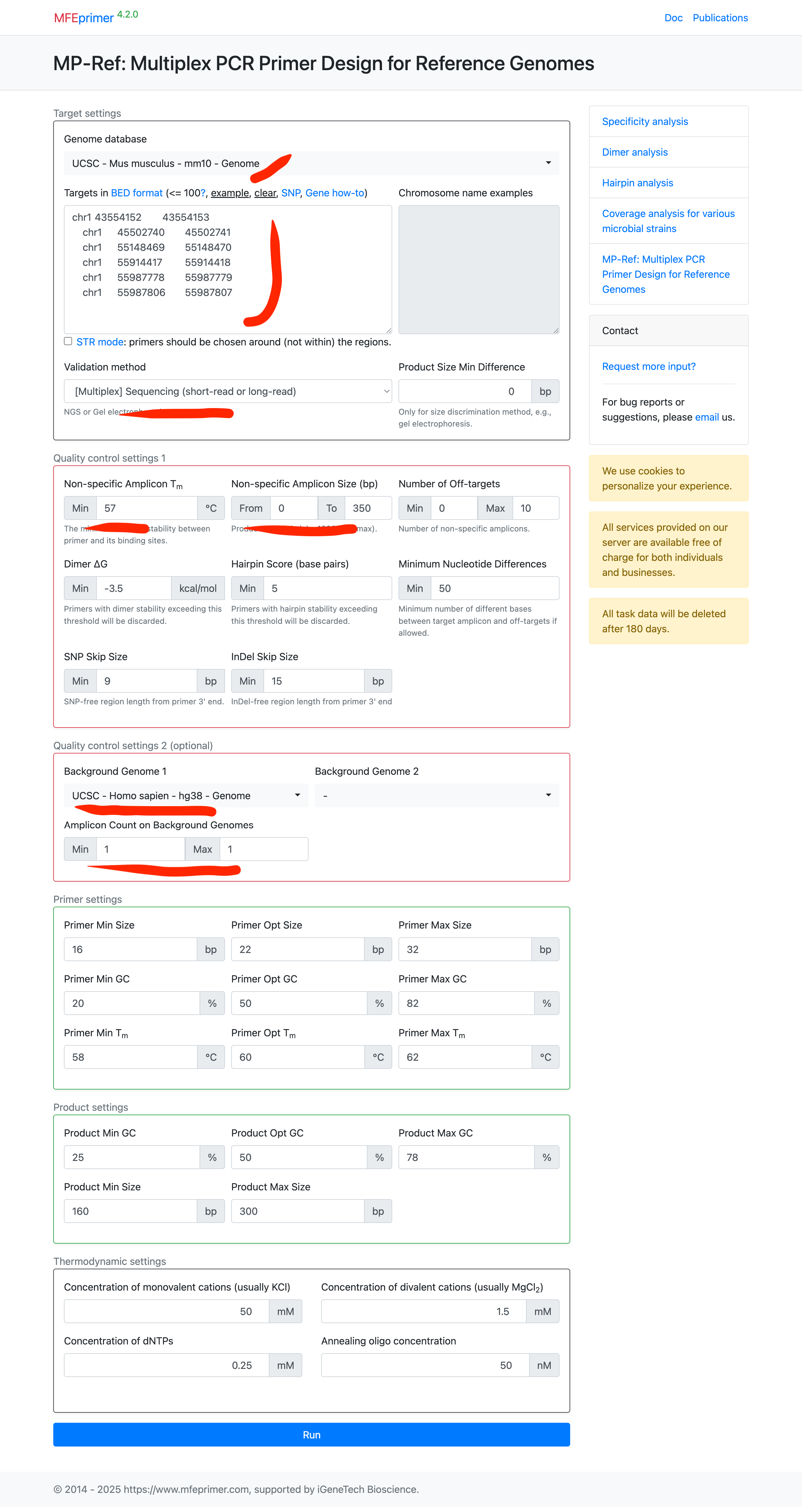
- The result page contains two parts, the first is “Coverage report”, listing all the target coverage in details. The second is “Primer list”, list all the primers in details. Each of them can be downloaded for further use. In this example, only one target can’t find a proper pair of primers to amplify both the mouse and human genome. And to be noted that, the last two targets is close enough that one pair of primers can amplify them both. So ther is only 4 pair of primers can amplify 5 targets.